

30 Minutes
Docker can build images based on the instructions from a Dockerfile. The instructions can be used for example to run arbitrary commands in the container and to copy files. Images can extend on existing images.
Create a new text file named Dockerfile in a temporarly directory.
cd /tmp/authorcarpentry-docker/
touch Dockerfile
Open it with a text editor and paste the following content
FROM rocker/rstudio
WORKDIR /home/rstudio
COPY articles.csv articles.csv
RUN install2.r wordcloud tm
We use Rocker’s RStudio as the base image and copy the file articles.csv into the container to the directory /data.
Now we must tell Docker to create an image.
docker build --tag authorcarpentry-docker-articles .
We use the tag authorcarpentry-docker-articles to be able to identify the image.
No start the image as usual and open the file in RStudio (go to http://localhost:8787 > File > Open File).
docker run -it -p 8787:8787 authorcarpentry-docker-articles
Now you can load and work with the data in R. Analysis example is based on https://www.r-bloggers.com/building-wordclouds-in-r/.
data <- read.csv("articles.csv")
head(data)
library(wordcloud); library(tm)
titles <- VCorpus(VectorSource(data$Title))
titles <- tm_map(titles, PlainTextDocument)
wordcloud(titles)
You can learn all about Dockerfiles in the Docker docs.
How is this useful for reproducible research?
You can create images tailored to your use case in a transparent way. If Docker every disappears, at least you will have all the instructions you originally used to create the image.
| Command | Explanation |
|---|---|
FROM |
specifies which base image, or stack of base images, your image is built on |
MAINTAINER |
specifies who created and maintains the image. |
CMD |
specifies the command to run immediately when a container is started from this image, unless you specify a different command. |
COPY |
will copy new files from a source and add them to the containers filesystem path |
WORKDIR |
sets the current directory for RUN and COPY statements (preferred over using RUN cd ..) |
RUN |
does just that: It runs a command inside the container (eg. apt-get) |
EXPOSE |
tells Docker that the container will listen on the specified port when it starts and the host may bind these |
VOLUME |
will create a mount point with the specified name and tell Docker that the volume may be mounted by the host |
Of course you can do more than just run an R script. You can also compile a literate programming document, such as a scientific publication.
Let’s start with this example R Markdown document:
---
title: "Magma Demo"
output: html_document
---
This example is based on http://rmarkdown.rstudio.com/lesson-3.html.
```{r include = FALSE}
knitr::opts_chunk$set(echo = FALSE)
```
```{r message = FALSE, warning = FALSE}
library(viridis)
```
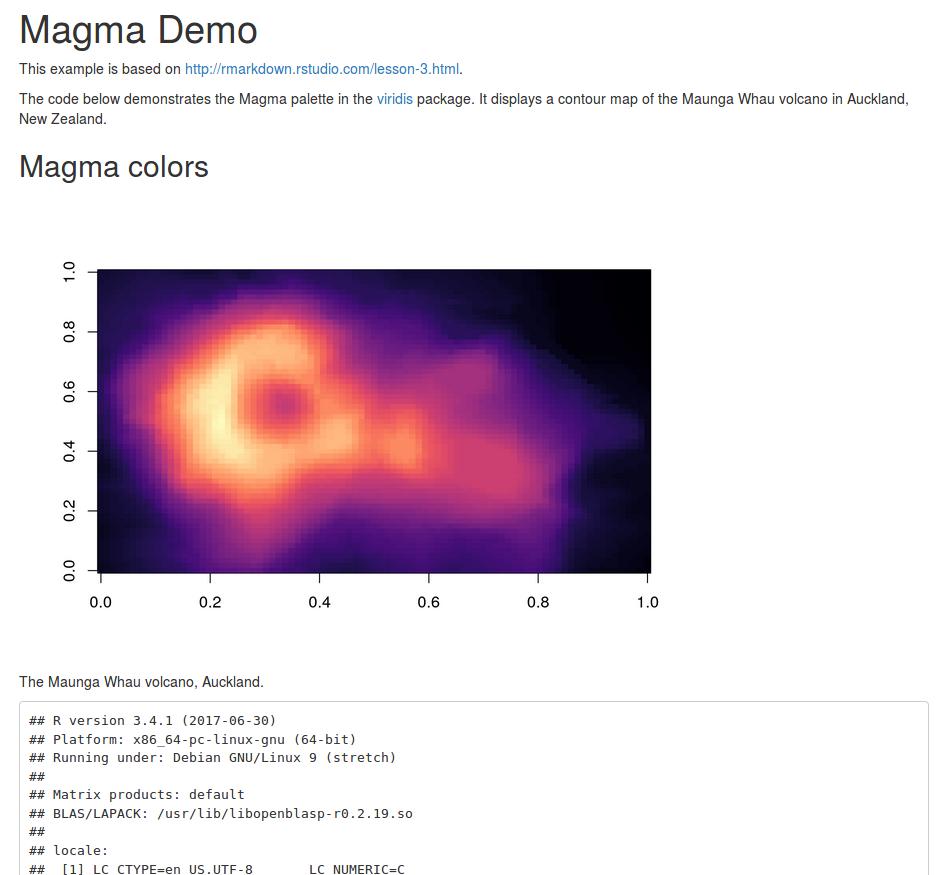
The code below demonstrates the Magma palette in the [viridis](https://github.com/sjmgarnier/viridis) package.
It displays a contour map of the Maunga Whau volcano in Auckland, New Zealand.
## Magma colors
```{r fig.cap = "The Maunga Whau volcano, Auckland."}
image(volcano, col = viridis(200, option = "A"))
```
```{r session}
sessionInfo()
```
Put this into a file paper.Rmd in a new directory on your computer.
The following Dockerfile starts from a version-fixed Rocker image, installs the required packages, and sets the rendering of the file as the image’s default command.
It also uses Label Schema labels to embed the required documentation into the image.
Put the content of the snippet below into a file Dockerfile next to paper.Rmd.
FROM rocker/r-ver:3.4.1
RUN apt-get update -qq \
&& apt-get install -y --no-install-recommends \
## Packages required by R extension packages
# required by rmarkdown:
lmodern \
pandoc \
&& apt-get clean \
&& rm -rf /var/lib/apt/lists/*
# install R extension packages
RUN install2.r -r "http://cran.rstudio.com" \
rmarkdown \
viridis \
&& rm -rf /tmp/downloaded_packages/ /tmp/*.rd
# add metadata
LABEL maintainer="Me! <mail@me.edu>" \
org.label-schema.usage="Run the image, then extract the created document with 'docker cp my-paper-container:/analysis/paper.html paper-from-container.html'" \
org.label-schema.docker.cmd="docker run -it --name my-paper-container imagename" \
org.label-schema.name="Docker for RR Demo" \
org.label-schema.schema-version="1.0"
WORKDIR /analysis
COPY paper.Rmd paper.Rmd
ENTRYPOINT ["sh", "-c"]
CMD ["R --vanilla -e \"rmarkdown::render(input = '/analysis/paper.Rmd', output_format = rmarkdown::html_document())\""]
Build the Dockerfile with docker build --tag my-paper .
Run the image with docker run -it --name my-paper-container my-paper and inspect the output on the console.
We name the container so that we can more easily retrieve files from it.
Extract the created output file with docker cp.
docker cp my-paper-container:/analysis/paper.html paper-from-container.html
You will see the following file:
Open the source code of the HTML document (Ctrl + U in most browses) and inspect it.
Noteworthy is the “embedding” of the image data in base64:
<img src="data:image/png;base64,iVBORw0KGgoAAAAN...
How is this relevant for research?
You can create a controlled environment to test your analysis. You will not miss any installed dependency on your computer which you are not conscious about, because the Dockerfile is a new environment. Archive the image, and you will be able to re-run the analysis anytime, even exchange data or render a different document using the same environment.